CryoEM Current Practices: Previous webinars

Some tips & tricks of note:
Sample preparation: Use of different detergents and grid types to find optimal ice thickness.
Data collection: Use of screening to inform decisions for data collection.
Data processing: Topaz picking to enrich views of a sub-population. Sorting damaged and intact particles. Heterogeneous refinement.
Complementary techniques: X-ray crystallography and comparison of structures solved by multiple techniques. Structure guided site directed mutagenesis to probe mechanistic hypothesis. Structure guided inhibitor design.
Some tips & tricks of note:
Sample preparation: Use of a wide screening space of different grid types to optimize sample behavior during plunge freezing. Use of graphene oxide films.
Data processing: Overcoming heterogeneity with multi-class ab initio and multi-class heterogeneous refinement. Decoy heterogeneous refinement.
Complementary techniques: Fluorescence polarization DNA binding studies for high throughput sample screening. Small angle X-ray scattering, and analytical ultracentrifugation to characterize oligomerizaton states.

Some tips & tricks of note:
Sample preparation: Purification of natively expressed proteins from C. elegans. Fluorescence Size Exclusion Chromatography (FSEC) for expression and purification screening. Use of thin carbon cryoEM grids for low-abundance samples.
Data processing: Heterogeneous refinement and analysis. Re-centering of particles to resolve distal ends of an elongated structure. Modeling of lipids into EM maps.
Complementary techniques: Use of AlphaFold predictions for structural analysis and model building. Mass spectrometry to characterize natively expressed protein complexes. Knockdowns in model organisms to validate protein complexes identified by cryoEM.

Some tips & tricks of note:
Sample preparation: Genetic covalent fusion to trap conformational states of a highly mobile molecular machine. Negative stain EM to troubleshoot sample dissociation. Buffer pH as an important variable for observed conformational states. Graphene oxide to prevent sample denaturation during vitrification.
Data collection: Targeting data collection on graphene oxide grids.
Data processing: Extensive 3D classification to resolve a dynamic sample. SAMUEL (Liao Lab tool). Focused classification & multibody refinement. “Off-centered” particle picking for focused refinement.
Complementary techniques: Correlation of cryoEM data to single molecule fluorescence studies.

Some tips & tricks of note:
Sample preparation: Optimization of protein-ligand incubation time for filament size.
Data processing: Manual and Topaz picking for filaments. Optimizing box size for filaments. Heterogeneous refinement for particle stack cleaning and improving orientation distribution. Comparison of reconstructions with different substrates/analogs to propose mechanisms.
Complementary techniques: SEC-MALS. Use of point mutations to validate interfaces observed in EM.
Recording coming soon

Some tips & tricks of note:
Data analysis: Building atomic models into cryoEM maps. Model validation tools.

Some tips & tricks of note:
Sample preparation: DNA mini-circles for structural studies. Use of support films/amorphous carbon. Optimizing glow discharge for thin carbon films.
Data collection: Tilted collection to overcome preferred orientation.
Data processing: Particle stack cleaning with “decoy classification”. 3D classification to identify multiple classes in a single data set.

Some tips & tricks of note:
Sample preparation: Membrane protein purification in detergent and detergent-CHS micelles. Purification of multiple orthologs.
Data collection: Hardware vs software binning to speed up data collection.
Data processing: Improving resolution with particle sorting in 3D.
Complementary techniques: Metal identification for modeling (Check My Metal server). Electrophysiology and SPR to characterize ligand/inhibitor binding. Use of structures determined with different ligands/inhibitors to determine mechanisms and classify antagonists

Some tips & tricks of note:
Sample preparation: In vitro production, purification, & refolding of RNAs. RNA specific considerations for grid prep. Use of stem extensions in RNA constructs to validate models.
Data processing: Model validation tools, especially for low resolution reconstructions. Processing of small and large RNAs.

Some tips & tricks of note:
Sample preparation: Graphene-oxide films to alleviate preferred orientation.
Data processing: 3D Variability analysis and 3D classification to understand inter-domain motion. Building a mechanistic model from multiple reconstructions.
Complementary techniques: SAXS for modeling and validation. Use of high-resolution X-ray structures to support ligand identification.

Some tips & tricks of note:
Sample preparation: Negative stain to characterize protein preps. Iterative screening to optimize grids. Importance of grid hole size. Use of toxins to stabilize structure.
Complementary techniques: Thermostability assays, X-ray crystallography, panning of synthetic Fabs for use as structural chaperones.
Other: Leveraging funding opportunities to travel for cryoEM training.

Some tips & tricks of note:
Sample preparation: Native and denaturing gel analysis of SEC fractions to use the most pure sample. Detergents as additives. Poly-lysine coated grids. Graphene vs graphene oxide grids to overcome aggregation and low particle density. Analysis of graphene oxide quality on grids.
Data processing: Separating conformational heterogeneity. 3D-variability analysis. Correlation of observed conformations to mechanistic steps.
Complementary techniques: DNA cleavage assays to characterize variants of interest. Kinetic modeling of assay data to generate mechanistic models.

Some tips & tricks of note:
Sample preparation: Mass photometry for sample characterization. Affect of different grid types.
Data processing: Kihara lab CryoEM software tools: EMSuite Server, careful particle curation, topaz picking.
Some tips & tricks of note:
Sample preparation: Mammalian cell expression. Expression of homo-oligomeric point variants. Negative stain for quality control. Graphene oxide grids.
Data processing: Multi-class ab-initio & non-uniform refinement to parse heterogeneity. Assigning lipid density in a transmembrane domain. Use of morphs between reconstructions to understand motion.
Complementary techniques: Electrophysiology

Some tips & tricks of note:
Sample preparation: Isolation of amyloids from tissue, negative staining for sample quality control.
Data processing: Manual picking for filament reconstruction. Separation of multiple classes from single filaments.
Complementary techniques: Mass spectrometry to verify composition of tissue extracted samples.

Some tips & tricks of note:
Sample preparation: tRNA scaffolds and Fab chaperones for RNA structure determination. Grid pre-treatments to increase particle density in grid holes. chameleon grid preparation.
Data processing: Multi-class ab initio to sort heterogeneity. Iterative workflows to obtain a best particle stack.
Complementary techniques: X-ray crystallography to obtain structures of sub-domains. SAXS analysis to verify folding.

Some tips & tricks of note:
Sample preparation: Negative stain EM to characterize oligomeric states.
Data collection: Minimizing dose during data collection to resolve flexible regions of a protein. Use of screening to ensure high quality data.
Data processing: Use of symmetry in 2D classification to understand binding stoichiometry.
Complementary techniques: Comparison to X-ray crystal structures to understand biophysical behavior and answer biological questions.
Other: Best use of local resources for impactful use of national center resources.
Some tips & tricks of note:
Sample preparation: NabFab strategy for particle picking and alignment of small membrane proteins. Nanodiscs for integral membrane proteins.
Data collection: Tilted data collection.
Data processing: Q-score to evaluate resolution. Processing a sample with heterogeneous conformations to find high-energy states.
Complementary techniques: HDX-MS & MD to characterize Nanobody (Nb) binding & conformational states. Nb characterization by two-hybrid assay. ITC to characterize NabFab and ligand binding to Nb bound transporter.
Some tips & tricks of note:
Sample preparation: Mixed expression systems to reconstitute complexes in vitro. Use of detergents to solve aggregation during plunge freezing.
Data processing: Topaz picking to obtain clean particle stacks. Focused refinement and composite maps for model building and refinement.
Complementary techniques: NMR to assign sequence registry in models when density is weak.
Some tips & tricks of note:
Data processing: Symmetry expansion and focused classification. Use of consensus maps for model building. Visualizing small ligand binding.
Complementary techniques: AI protein design, selection of target binding DARPins.

Some tips & tricks of note:
Sample preparation: Tag locations on heterologously expressed protein constructs. Time resolved cryo-EM from on-grid enzymatic reactions.
Data processing: Handling heterogeneous oligimerization. 3D Variability Analysis (3DVA). Ensemble generation from 3DVA. Assigning small ligand densities.
Complementary techniques: Crosslinking to validate oligomerization interfaces. Enzyme kinetics. Molecular Dynamics Ensemble Refinement. Using cryoEM results to drive development of Undergraduate CURE lab courses.

Some tips & tricks of note:
Sample preparation: Tips for dealing with low sample concentration, samples that stick to carbon, recognizing and troubleshooting strange types of ice.
Data collection: Using information from different magnifications to characterize grids and optimize data collection.
Data processing: How to do gain correction when you don’t have gain references. Helical processing challenges.

Tips & tricks of note:
Sample preparation: membrane protein purification using GDN & nano-disks, expression of leaky ion channels.
Data processing: multi-software workflows, masking and local refinement, symmetry expansion, variability analysis.
Complimentary techniques: patch clamping, single amino acid substitution to verify binding sites.
Tips & tricks of note (coming soon):
Sample preparation:
Data processing:
Complimentary techniques:
Tips & tricks of note (coming soon):
Sample preparation:
Data processing:
Complimentary techniques:
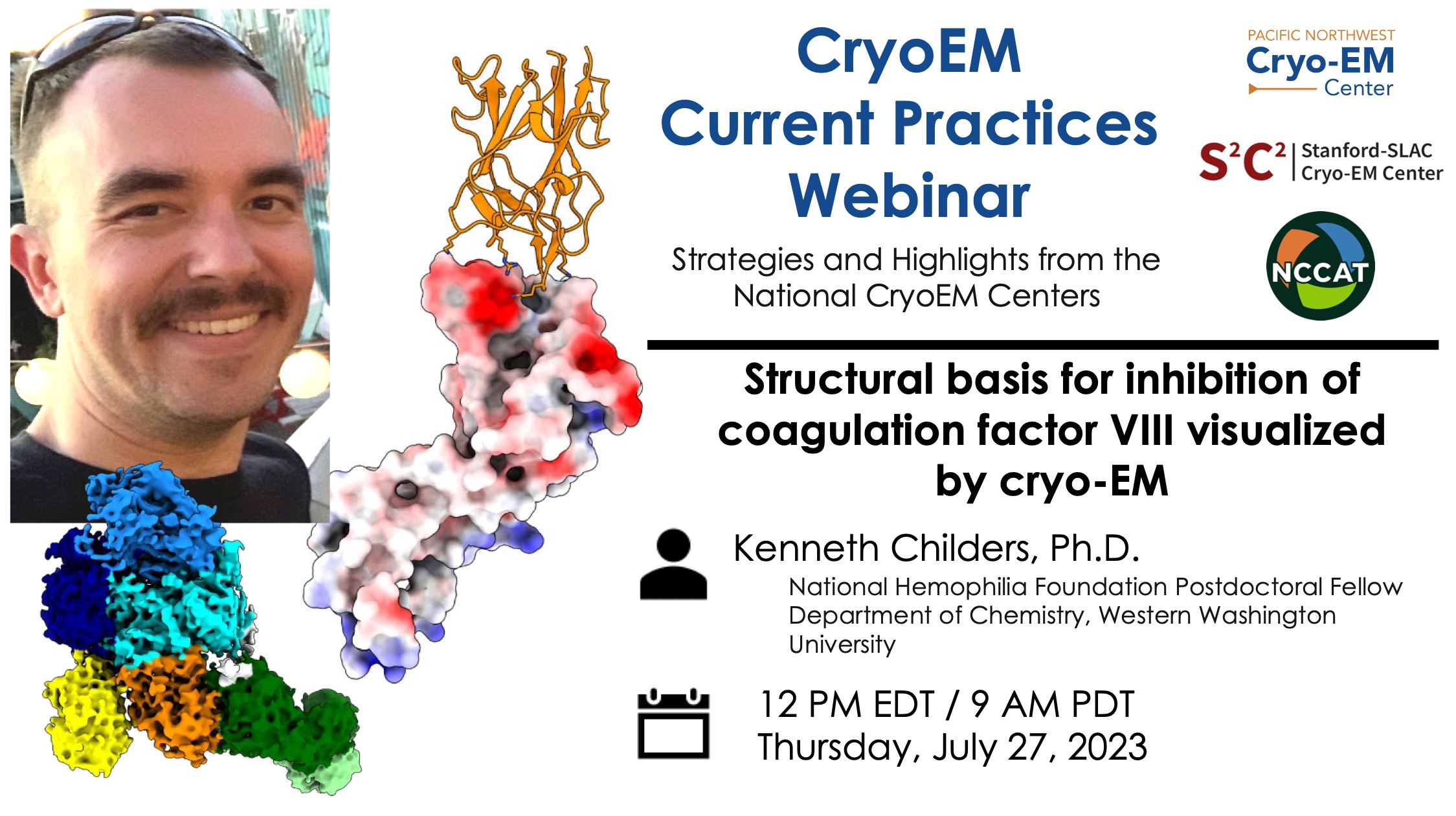
Tips & tricks of note:
Sample preparation: Continuous thin carbon substrates to reduce aggregation.
Data processing: Local refinement to improve map quality in regions of interest.
Complimentary techniques:

Tips & tricks of note:

Tips & tricks of note:
Sample preparation: Anaerobic Grid preparation.
Data processing: “In silico purification” of heterogeneous sample (enzyme in turnover conditions). Determination of multiple structures from one grid. Local resolution analysis to understand dyamics. High resolution to analyze single amino acid side-chain movement.
Complimentary techniques: Protein design. Metal-mediated protein oligomerization to generate extended molecular assemblies. Analytical ultracentrifugation. Electron diffraction.
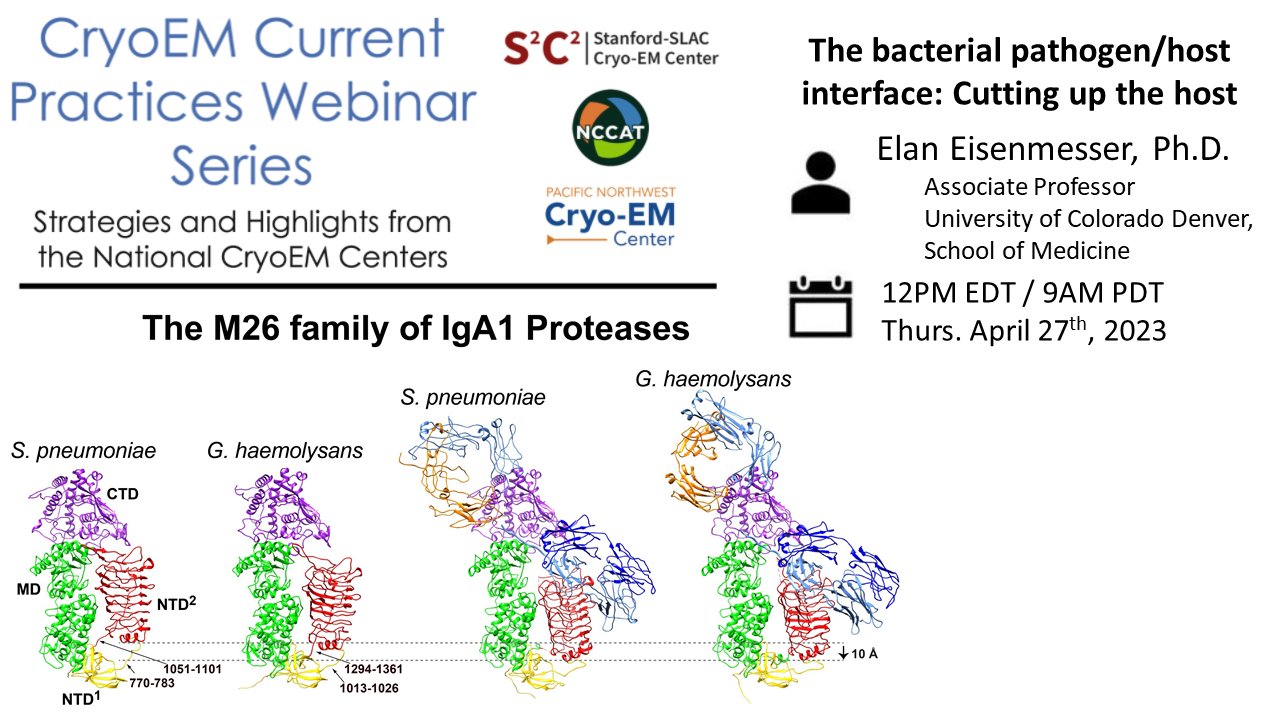
Tips & tricks of note:
Sample preparation: Using NMR analysis of point-mutants to choose an ideal cryoEM sample. Ideas for overcoming preferred orientation.
Data processing: Understanding substrate specificity by solving multiple structures.
Alternative methods: Site-directed mutagenesis, NMR as a complementary technique, RMSD analysis of substrate bound/unbound structures.
Tips & tricks of note:
Sample preparation: Blot-free vitrification to overcome preferred orientations
Data collection: Tilted data collection to overcome preferred orientations
Data processing: Using neural network particle pickers to find harder-to-pick views of non-spherical particles
Tips & tricks of note:
Sample preparation: Use of C. elegans for native source expression of a mechanosensing protein. Purification of samples from native sources (including tag choice). Fluorescence-detection size-exclusion chromatography (FSEC) to optimize buffer conditions. Continuous-carbon support to increase sample concentration for precious samples. Amylamine treatment of cryoEM grids to alleviate preferred orientation and improve ice thickness.
Data processing: Heterogeneous refinement to reconstruct multiple conformations from a single data set. Masked local refinement to improve resolution a weak binding partner. Observation of phospholipids in reconstrution.
Complementary techniques: Mass spectrometry to characterize sample composition. Molecular dynamics to probe role of membrane deformation in mechanism.
Tips & tricks of note:
Data processing: Practicalities of neural network picker software installation and usage. How to choose a particle picker & validation. Neural network picking for single particle analysis, filaments, & tomography. Interplay of particle picking and preferred orientations. Particle centering.

Tips & tricks of note:
Sample preparation: Synthetic G-quadruplex design. Screening of different grid types. Micrograph evaluation for a small (sub-30 kDa) particle.
Data processing: Elliptical blob picking. Heterogenenous refinement and 3D variability to separate conformational heterogeneity. Non-uniform refinement.
Complementary techniques: Analytical Ultracentrifugation. 1H-NMR. Small angle X-ray Scattering (SAXS) for size and flexibility estimation. Molecular dynamics with cryoEM map derived restraints. Molecular dynamics of a cryoEM derived model. Computational drugability analysis.

Tips & tricks of note:
Sample preparation: Transmembrane proteins, HEK293 freestyle cell expression, purification using FLAG-tag, pH-dependent enzyme activity, thermal denaturation, golgi apparatus, muscular dystrophy, IEX, SEC-MALS, SAXS
Data processing: 2-D classification, template-based picking, unbinned repicking to eliminate bias and improve resolution, preferred orientation.

Tips & tricks of note:
Sample preparation: Protein purification from red blood cells using glycerol gradients & gel filtration, detergent solubilization & purification, detergent screening, digitoxin solubilization, mass photometry.
Data collection: Single particle analysis & Cryo-ET
Data processing: Particle heterogeneity, non-uniform refinement, class-based processing. Tomogram 2-D slices, Sub-tomgram averaging.

Tips & tricks of note:
Sample preparation: Fibril extraction from heart tissue, mutation-specific perturbations, amyloid seeding, use of structure-based inhibitors.
Data processing: Manual picking, conformational heterogeneity, local resolution analysis, featureless cylinder, helical processing, twist and local maxima.
Complimentary techniques: X-ray microcrystallography.

Tips & tricks of note:
Sample preparation: Screening of: detergents, nano disc size, temperatures, grid type, and plunge freezer type. Use of pre-heated tools for temperature sensitive samples.






Tips & tricks of note:
Sample preparation: Protein prep and grid freezing optimization, glutaraldehyde cross-linking
Data collection: Tilted data collection to overcome preferred orientations
Data processing: Tips for processing tilted data, particle picking tips for tricky particles, helical filament processing
A copy of this presentation is available on the Open Science Framework (OSF) repository: https://osf.io/x7dv8/

Tips & tricks of note:
Data processing: computational infrastructure and resources
Tips & tricks of note:
Sample preparation: Optimization of grid freezing conditions
Data processing: Tips for dynamic membrane proteins
Tips & tricks of note:
Sample preparation: Testing detergents and nanodisks for integral membrane proteins.
Data collection: Optimizing pixel size and data collection rate.
Data processing: Using symmetry during 3D reconstruction; cleaning particle stacks with 3D classification.

Tips & tricks of note:
Sample preparation: Sample QC using differential scanning calorimetry & negative stain EM. The effects of temperature on protein quality and grid preparation.
